Note: If they do not exist already, these values must be added to the headers of your .fits files in your Telescope Control System when writing the files. Many of them can be pulled out of the Observation Portal’s schedule observation json and put directly into the header of data taken for that observation.
Observatory Data Flow
For users who wish to utilize the Science Archive for data archival and retrieval, this document describes the components necessary.
Main Components
- OCS Ingester - commonly used as a library to simplify the ingestion process of observatory data products into the science archive.
- OCS Science Archive - A Django application which aids in the archival storage and retrieval of observatory data products.
Both of these applications make use of a common library, the OCS Archive Library. The OCS Archive library allows the OCS Ingester and Science Archive to share common configuration and reduces the need for duplicated code across the two projects - this shared code contains classes used to abstract data files and data storage backends. See the Advanced Topics section for more information about customizing the behavior of the ocs_archive library.
Configuration
Because both the OCS Science Archive and OCS Ingester depend on the OCS Archive Library, the two can share common configuration. Runtime configuration for the OCS Archive Library is set using environment variables - see OCS Archive Configuration Documentation for more detail about possible configuration options.
Setting up FITS Headers
The OCS Archive Library has built in capacity to handle .fits files, but certain environment variables must be set to map between .fits header keys and concepts that are required by the Science Archive for data filtering. The table below lists the .fits header concepts expected, their environment variable to override the header key, and the default header key.
| Archive model field | Description | Environment Variable | Default Header Key |
|---|---|---|---|
| observation_date | The time of the observation in UTC | OBSERVATION_DATE_KEY | DATE-OBS |
| observation_day | The observing night in YYYYMMDD | OBSERVATION_DAY_KEY | DAY-OBS |
| site_id | The configdb site code the data was taken at | SITE_ID_KEY | SITEID |
| telescope_id | The configdb telescope code this data was taken at | TELESCOPE_ID_KEY | TELID |
| instrument_id | The configdb instrument code this data was taken with | INSTRUMENT_ID_KEY | INSTRUME |
| observation_id | The observation ID for this piece of data | OBSERVATION_ID_KEY | BLKUID |
| request_id | The request ID for this piece of data | REQUEST_ID_KEY | REQNUM |
| target_name | The user defined name of the target for this data | TARGET_NAME_KEY | OBJECT |
| configuration_type | The configuration type of this observation | CONFIGURATION_TYPE_KEY | OBSTYPE |
| proposal_id | The proposal ID this data was requested under | PROPOSAL_ID_KEY | PROPID |
| public_date | The date at which this data should be made publicly accessible | PUBLIC_DATE_KEY | L1PUBDAT |
| primary_optical_element | The value of the primary optical element this data was taken with | PRIMARY_OPTICAL_ELEMENT_KEY | FILTER |
| exposure_time | The exposure time this data was taken with | EXPOSURE_TIME_KEY | EXPTIME |
| reduction level | The reduction level of this data (0 is considered raw). | REDUCTION_LEVEL_KEY | RLEVEL |
There are also several environment variables for mapping .fits header keys that are used in the ingestion of data but not stored directly into the archive. These include:
| Environment Variable | Description | Default Header Key |
|---|---|---|
| OBSERVATION_END_TIME_KEY | The ISO formatted observation end date | UTSTOP |
| CONFIGURATION_ID_KEY | The configuration ID for this piece of data | MOLUID |
| REQUESTGROUP_ID_KEY | The requestgroup ID for this piece of data | TRACKNUM |
| CATALOG_TARGET_FRAME_KEY | The base filename of the catalog file for this target | L1IDCAT |
| RADIUS_KEY | The FOV radius in arcseconds for a circular FOV, used to calculate WCS if specified | RADIUS |
| RA_KEY | The FOV center RA for a circular FOV, used to calculate WCS if specified | RA |
| DEC_KEY | The FOV center DEC for a circular FOV, used to calculate WCS if specified | DEC |
| RELATED_FRAME_KEYS | A comma delimited list of header keys to search for related frame base filenames in the header | L1IDBIAS,L1IDDARK,L1IDFLAT,L1IDSHUT,L1IDMASK,L1IDFRNG,L1IDCAT,L1IDARC,L1ID1D,L1ID2D,L1IDSUM,TARFILE,ORIGNAME,ARCFILE,FLATFILE,GUIDETAR |
If there are more complicated mappings that you need to pull the data out of your .fits file, or if you have a different file type entirely, then please read the section below on adding custom dataproduct classes.
Telescope Control System Integration
In the execution of an observation, the observatory’s Telescope Control System generates data products. For these products to be archived and served to users or staff via an OCS Science Archive, they must be ingested.
A Simple Ingestion Scheme
Once a TCS has created its final data products to be archived, the TCS may call out to the Ingester library directly. If the TCS is written in Python, the call is as simple as setting the relevant environment variables for your filestore type and running:
from ocs_ingester.ingester import upload_file_and_ingest_to_archive
with open('/path/to/file', 'rb') as file:
ingester_response = upload_file_and_ingest_to_archive(file)
...
# optionally do something with the ingester response
The Ingester Response contains information about the archived frame, such as its location (URL for cloud filestore), ID in the Science Archive database and other frame metadata. This can be useful if the application ingesting the data wishes to keep its own record of data products and their location in the archive.
For more detailed information about the OCS Ingester and its capabilities, see the developer documentation.
Advanced Topics
Overriding Classes in the ocs_archive Library
The ocs_archive library provides base classes to represent observatory data products and filestore locations in order to provide a unified interface to various types of data products and filestore solutions.
Data Products
Within the input module of the ocs_archive library there exist classes to represent various different types of files such as FITS, GZipped TAR files, etc…
If an observatory wishes to add their own file specification, they may do so by sub-classing the DataFile class and adding any customizations needed. For examples of this functionality, see the implementation of the FitsFile class, which customizes the base DataFile class to work with FITS files. For Las Cumbres Observatory’s purposes, we have further sub-classed the FitsFile class into an LCOFitsFile to add additional functionality to meet the needs of our observatory.
Once a new file specification has been added, it should be added to the FileFactory class, which is responsible for returning the appropriate DataFile subclass given a file’s extension. It utilizes a mapping from file extension -> DataFile class to determine which class to construct, given an input file:
EXTENSION_TO_FILE_CLASS = {
'.fits.fz': 'ocs_archive.input.lcofitsfile.LcoFitsFile',
'.fits': 'ocs_archive.input.fitsfile.FitsFile',
'.tar.gz': 'ocs_archive.input.tarwithfitsfile.TarWithFitsFile',
'.pdf': 'ocs_archive.input.file.DataFile'
}
This mapping can be overridden using the FILETYPE_MAPPING_OVERRIDES environment variable if the default is not sufficient or needs to be changed.
Filestore
A set of classes which represent storage backends exists within the storage module of the ocs_archive library.
The base FileStore class is intended to represent a data product storage backend, and can be sub-classed and extended to work with many different storage solutions.
For example, the S3Store contains customizations to the base FileStore class to allow observatory data products to be stored in Amazon S3. If an observatory needs to use a different cloud provider, then a new subclass can be created.
The ocs_archive also provides a FileStore class, FileSystemStore, made to work with network or local filestores, if an observatory does not wish to store their data with a cloud provider. Filesystem storage does not provide version history or time authenticated generated urls, so S3 Storage should be heavily preferred for production setups.
Persistent Queue for Data Products
Note: This only applies when a separate process or application is responsible for ingesting data products from the TCS
When a TCS generates data products to be ingested, it’s helpful to use a queueing system such as RabbitMQ to keep track of images to be ingested. By utilizing a simple queuing architecture, if the application responsible for ingesting images ever experiences downtime, the TCS can still queue images to be ingested and the ingestion application will be able to catch up after its outage.
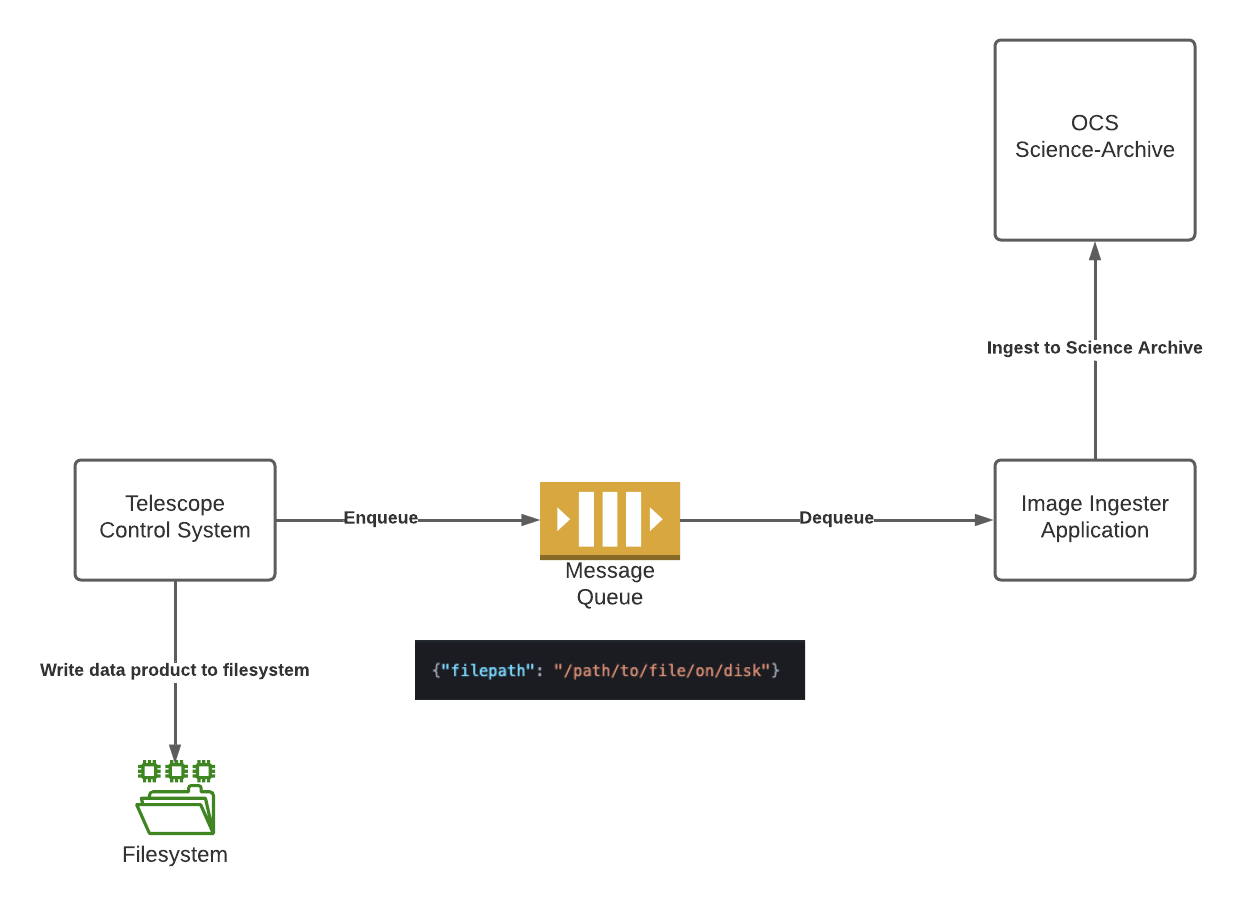
The queue message can be as simple as the location from which to retrieve the file to be ingested, but should contain the minimum amount of information necessary for the ingestion application to successfully ingest the data to the science archive.
{"filepath": "/path/to/file/on/disk"}
Retry Scheme for Data Product Ingestion
For added robustness, it is good practice to integrate retry logic into the data product ingestion process. It’s for this reason that the OCS Ingester defines a set of Python exception classes that will be raised in appropriate situations:
- RetryError - Exception that is raised when an error happens that can be retried.
- BackoffRetryError - Exception that is raised when an error happens that can be retried with an expontential backoff. For example, networking latency errors that may succeed at a later time.
- DoNotRetryError - Exception that is raised when an error happens that will undoubtedly repeat if called again. The task should not be retried.
- NonFatalDoNotRetryError - Exception that is raised when an error happens that should not be retried and is also not a fatal condition. This is different from the DoNotRetryError as it indicates the failure wasn’t a fatal issue in the ingestion of an image (e.g. file doesn’t exist on disk or Astropy exception raised), but rather indicates an error such as the file already existing in the filestore.
Handling these errors and retrying ingestion if appropriate will increase the robustness of an observatory’s data product flow. Appropriate retry logic combined with queueing infrastructure leads to a system which is fault-tolerant and able to recover easily from intermittent service outages.
In Python, the Tenacity package provides a set of function decorators that allow for various retry schemes.
Keeping a Record of Data Product Ingestion
Applications may need to keep track of some of the data products they’ve ingested into the Science Archive. For instance, a data pipeline that creates calibration frames for use in later science data reduction may need to access these images from the science archive to perform scientific reductions.
One method to keep a record of these data products is using a relational database table that the application can access as needed. When a data product is created by the application, it creates a new image record in its database containing the Science Archive frame ID alongside any other useful metadata that the application may need (filter used, binning, instrument name, etc…).
By storing the frame ID in the record, the lookup process in the Science Archive is greatly sped up, as the Science Archive record for this image can be accessed by simply by sending an HTTP GET request to the /frames/<image-ID> endpoint.
For an observatory whose Science Archive is handling many requests, this scheme prevents internal applications from adding unnecessary load to the system by allowing for near-instant access by ID, rather than using more expensive HTTP filter parameters.